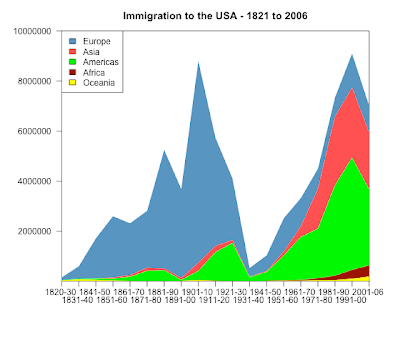
The data are c & p from here:
>original
Europe Asia Americas Africa Oceania
1820-30 106487 36 11951 17 33333
1831-40 495681 53 33424 54 69911
1841-50 1597442 141 62469 55 53144
1851-60 2452577 41538 74720 210 29169
1861-70 2065141 64759 166607 312 18005
1871-80 2271925 124160 404044 358 11704
1881-90 4735484 69942 426967 857 13363
1891-00 3555352 74862 38972 350 18028
1901-10 8056040 323543 361888 7368 46547
1911-20 4321887 247236 1143671 8443 14574
1921-30 2463194 112059 1516716 6286 8954
1931-40 347566 16595 160037 1750 2483
1941-50 621147 37028 354804 7367 14693
1951-60 1325727 153249 996944 14092 25467
1961-70 1123492 427642 1716374 28954 25215
1971-80 800368 1588178 1982735 80779 41254
1981-90 761550 2738157 3615225 176893 46237
1991-00 1359737 2795672 4486806 354939 98263
2001-06 1073726 2265696 3037122 446792 185986png("immigration_log_scatter_BW.png", width = 560, height = 480)
par( mar=c(7, 7, 3, 3) )
plot( original$Europe, log="y", type="l", col="grey20", lty=1,
ylim=c(10, 10000000), xlab="Year Interval", ylab="Number of Immigrants Admitted to the United States",
lwd=2, xaxt='n', yaxt='n', mgp=c(4.5,1,0) ) # xaxt='n' an d yaxt='n'- do not show x and y axis
for (i in 2:dim(original)[[2]]){
lines(original[, i], type="l", lty=i, col="grey20")
}
axis(1, 1:dim(original)[[1]], rownames(original), las=2)
axis(2, at=c(10,100,1000,10000,100000,1000000,10000000), labels=c(10,100,1000,10000,100000,1000000,10000000), las=2, tck=1, col="grey85")
box()
legend( 14,400, legend=colnames(original), lty=c(1:5) )
dev.off()png("immigration_stacked_chart.png", width = 560, height = 480)
library(plotrix)
par( mar=c(6, 6, 3, 3) , las=1)
colori4<-c("yellow", "darkred","green","brown1", "steelblue")
stackpoly( original[, 5:1], col=smoothColors(colori4), border=NA,stack=T, xaxlab=rownames(original),
ylim=c(10,10000000), staxx=TRUE, axis4=F, main="Immigration to the USA - 1821 to 2006" )
legend("topleft", legend=colnames(original), fill=smoothColors(colori4)[5:1] )
dev.off()